(Created page with "<gallery> File:20230412_105730.jpg File:20230412_111428.jpg File:20230412_143503.jpg File:20230412_150047.jpg File:20230412_150053.jpg File:20230412_150130.jpg File:20230412_1...") |
Lea.maravic (talk | contribs) (new photo) |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<gallery> | '''WORKSHOP I''' | ||
12.04.2023, 13.04.2023 - [[GMU:BioArt Forum/Growing, Shaping and Living (with) Microalgae|/Growing, Shaping and Living (with) Microalgae]] with biologist Johann Bauerfeind<gallery> | |||
File:20230412_105730.jpg | File:20230412_105730.jpg | ||
File:20230412_111428.jpg | File:20230412_111428.jpg | ||
| Line 29: | Line 31: | ||
File:20230417_110858.jpg | File:20230417_110858.jpg | ||
File:20230418_171027.jpg | File:20230418_171027.jpg | ||
</gallery> | </gallery>'''WORKSHOP II.''' | ||
28.04.2023, 29.04.2023 - [[GMU:BioArt Forum/DNA analysis|/DNA analysis]] with molecular biologist Julian Chollet | |||
Julian is a molecular biologist who showed us how to use a diagnostic tool called PCR (Polymerase Chain Reaction) for DNA analysis. All living beings contain a complex molecule called DNA polymerase. It copies DNA so all organisms that want to replicate have it. In order to replicate and or analyse a DNA in a laboratory we need 3 substances: DNA or an organism we want to analyse, DNA polymerase and primers. Primers are small pieces of DNA made up in a laboratory and they can have any sequence of nucleotides. | |||
For the experiment all the participants chose a substance to analyse and then we prepared the specimens. Mostly we worked with fruit. Some of us chose saliva as our specimen. Most specimens were prepared by first chopping, blending, mushing and centrifuging them. Then we put them through 3 stages: | |||
1) DENATURATION - heating to 95°C to break up the connections between DNA helyx | |||
2) ANNEALING - at the temperature of 55°C primers will attach to the basis of DNA | |||
3) EXTENSION - the temperature of 72°C is the point when DNA polymerase replicates DNA by extending the primer. It assembles 1000 pieces of DNA at this temperature | |||
[[File:20230428 134426.jpg|thumb|1082x1082px|Preparing samples]] | |||
After the preparation was finished, we loaded these samples in one neat row on a gel bed connected to electrodes. After half an hour the florescent light showed that she samples assembled themselves in columns at various lengths. We used the raspberry as our control and the raspberry sample columns all assembled themselves to the same hight as the control. We could determine which samples are of plant and which of animal origin. Very few samples were successfully put through this PCR test. | |||
[[File:20230428 134817.jpg|thumb|1082x1082px|Preparing samples 2[[File:Results.jpg|thumb|1548x1548px|Results]]]] | |||
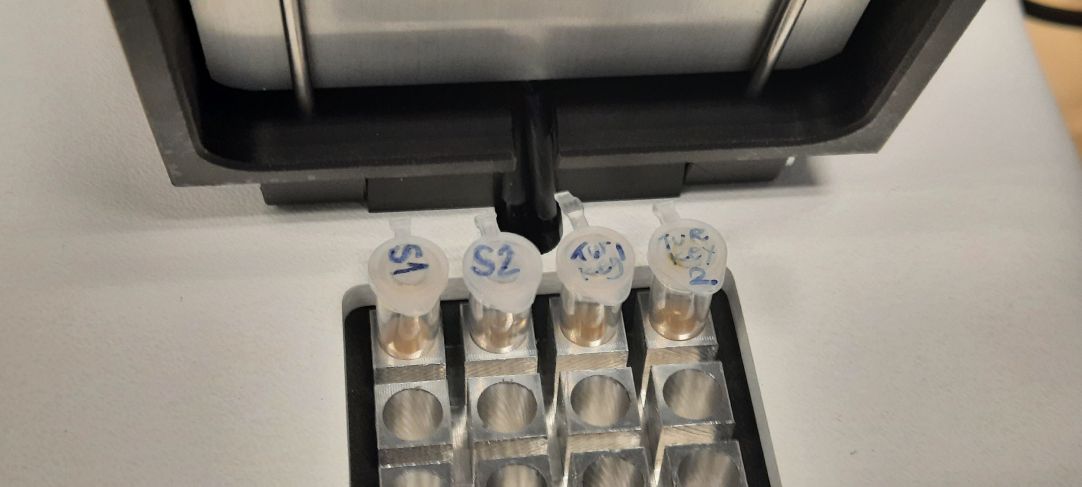
[[File:20230521 115319.jpg|thumb|341x341px|Setting up technology[[File:Nanopore active cells.jpg|thumb|345x345px|Nanopore active cells]]]] | |||
'''WORKSHOP III.''' | |||
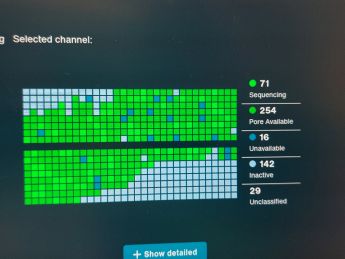
20.05.2023, 21.05.2023 - [[GMU:BioArt Forum/Nanopore sequencing|/Nanopore sequencing]] with safety engineer Lisa Thalheim[[File:MinION sequencer with flow cell.jpg|thumb|340x340px|MinION sequencer with flow cell that is being loaded with the sample[[File:Comparing the results with online libraries.jpg|thumb|345x345px|Comparing the results with online libraries]]]]Lisa gave us a beautiful overview of an Artist's Guide to DNA Sequencing and how the technology was previously used by artists. After the introduction we were acquainted with the Oxford Nanopore Technologies (https://nanoporetech.com/) and the equipment we later used for DNA sequencing. | |||
Once in the laboratory we extracted small amounts of the fluid from multiple cyanobacteria and algae cultures. We then prepared the sample by adding various buffers according to the prescribed protocol. | |||
The sample contained many organisms and once the sequencing was done we could compare the results to the information in online libraries and identify the organisms present in our sample. | |||
Latest revision as of 19:31, 21 July 2023
WORKSHOP I
12.04.2023, 13.04.2023 - /Growing, Shaping and Living (with) Microalgae with biologist Johann Bauerfeind
WORKSHOP II.
28.04.2023, 29.04.2023 - /DNA analysis with molecular biologist Julian Chollet
Julian is a molecular biologist who showed us how to use a diagnostic tool called PCR (Polymerase Chain Reaction) for DNA analysis. All living beings contain a complex molecule called DNA polymerase. It copies DNA so all organisms that want to replicate have it. In order to replicate and or analyse a DNA in a laboratory we need 3 substances: DNA or an organism we want to analyse, DNA polymerase and primers. Primers are small pieces of DNA made up in a laboratory and they can have any sequence of nucleotides.
For the experiment all the participants chose a substance to analyse and then we prepared the specimens. Mostly we worked with fruit. Some of us chose saliva as our specimen. Most specimens were prepared by first chopping, blending, mushing and centrifuging them. Then we put them through 3 stages:
1) DENATURATION - heating to 95°C to break up the connections between DNA helyx
2) ANNEALING - at the temperature of 55°C primers will attach to the basis of DNA
3) EXTENSION - the temperature of 72°C is the point when DNA polymerase replicates DNA by extending the primer. It assembles 1000 pieces of DNA at this temperature
After the preparation was finished, we loaded these samples in one neat row on a gel bed connected to electrodes. After half an hour the florescent light showed that she samples assembled themselves in columns at various lengths. We used the raspberry as our control and the raspberry sample columns all assembled themselves to the same hight as the control. We could determine which samples are of plant and which of animal origin. Very few samples were successfully put through this PCR test.
WORKSHOP III.
20.05.2023, 21.05.2023 - /Nanopore sequencing with safety engineer Lisa Thalheim
Lisa gave us a beautiful overview of an Artist's Guide to DNA Sequencing and how the technology was previously used by artists. After the introduction we were acquainted with the Oxford Nanopore Technologies (https://nanoporetech.com/) and the equipment we later used for DNA sequencing.
Once in the laboratory we extracted small amounts of the fluid from multiple cyanobacteria and algae cultures. We then prepared the sample by adding various buffers according to the prescribed protocol.
The sample contained many organisms and once the sequencing was done we could compare the results to the information in online libraries and identify the organisms present in our sample.